|
Goal: Compare the average volume of the major protrusion for untreated cells and CK666 treated cells.
Got all the major protrusion volumes untreated.largest.prot = c(c14_protrusions_rv2$V3, c15_protrusions_rv$V3, c10_protrusions_rv2$V3, c12_protrusions_rv$V3) Found the mean and the standard dev mean(untreated.largest.prot) sd(untreated.largest.prot) Note: for setting all NAs to 0: x[is.na(x)] <- 0 Results: The average volume of the major protrusion is higher, but also has a very large standard deviation.
0 Comments
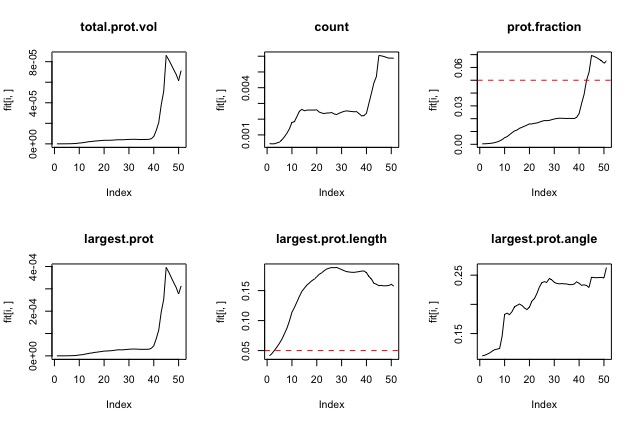
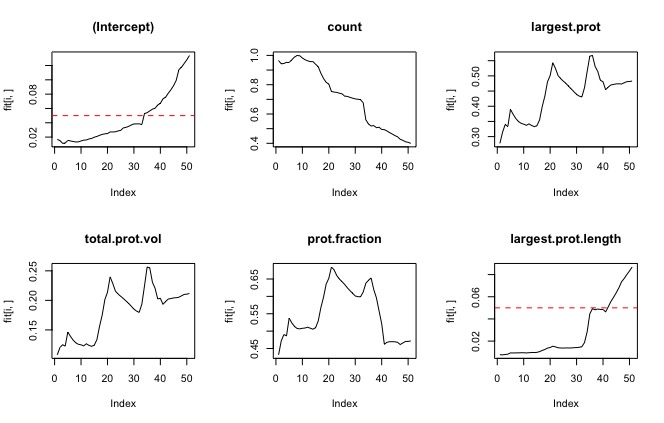
Goal: To see if there is a correlation between the protrusion parameters matrix and the math of the cells using linear regression. Training matrices: DMSO treated cells (in order): cell 14, cell 15, cell 12, cell 10 CK666 treated cells (in order): cell 43 ,cell 48 , cell 55 , cell 53 variables: Count, largest protusion volume, total protustion volume, protrusion fraction, largest protrusion length, largest protrusion angle, and total protrusion angle. Experiment 1Dependent value (y): derivative using savitzky-golay variables:
To get rid of rows without Y values training.set.complete<-training.set[complete.cases(training.set),] I did this because the smoothing window on the sg filter leaves a lot of blank values for the derivative. Results: all the parameters were significant except largest protrusion angle, and total angles. The largest protrusion length is only significant for the first few fits. Experiment 2Dependent value (y): curve method variables:
Experiment 3Dependent value (y): bi-modal with cutoff (ie, is the cell turning or not turning?)
Results:
To do next:
add more cells to the training set write function to find average turning point prediction |
Categories
All
|
 RSS Feed
RSS Feed