|
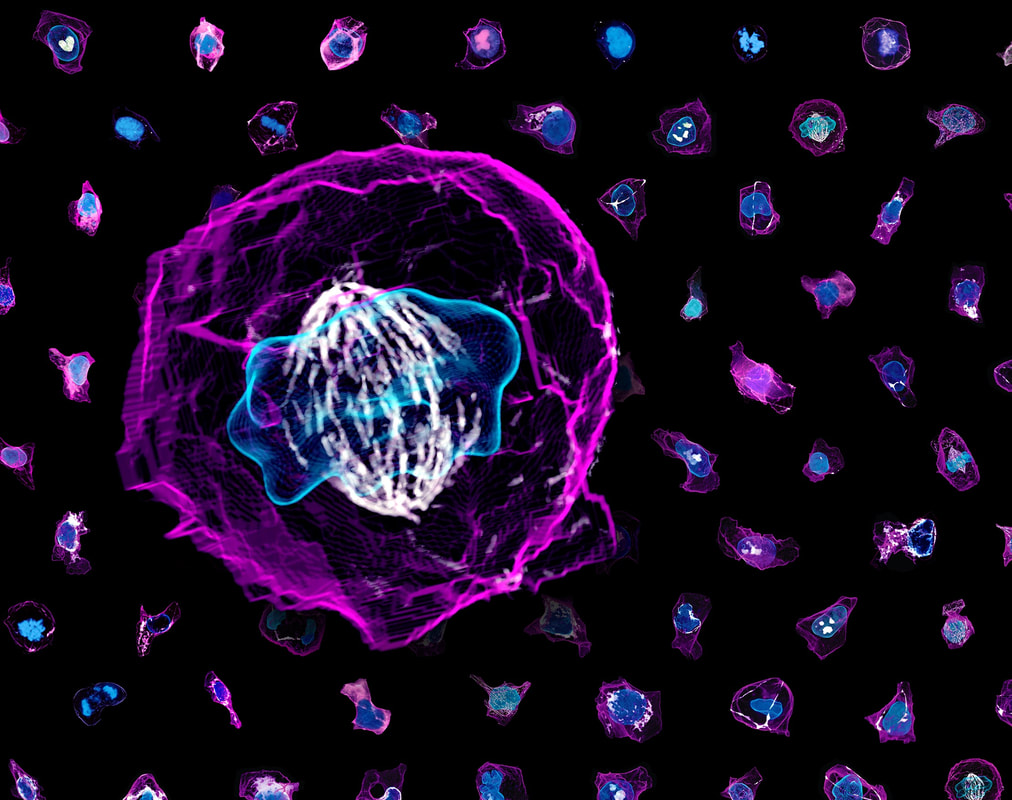
Integrated intracellular organization and its variations in human iPS cells
Database of 200,000 cell images yields new mathematical framework to understand our cellular building blocks. Link to write up about the article.
0 Comments
The Simularium Viewer: an interactive online tool for sharing spatiotemporal biological models.
Blair Lyons, Eric Isaac, Na Hyung Choi, Thao P. Do, Justin Domingus, Janet Iwasa, Andrew Leonard, Megan Riel-Mehan, Emily Rodgers, Lisa Schaefbauer, Daniel Toloudis, Olivia Waltner, Lyndsay Wilhelm & Graham T. Johnson. Nature Methods (2022). https://doi.org/10.1038/s41592-022-01442-1
I worked in collaboration with Daniel Toloudis under the supervision of Graham Johnson, to build the 3D cell viewer at allencell.org. We are currently working on version 2.0
As part of a project to feature research at UCSF, my collaborator, Lilllian Fritz-Laylin and I were interviewed about our research.
Traditional illustration techniques and computational approaches to visualize and analyze state of the art microscopy data
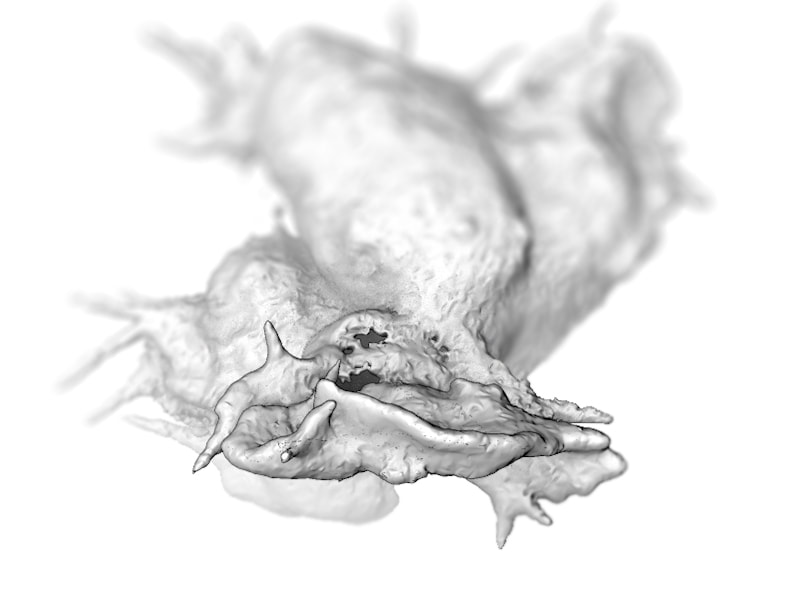
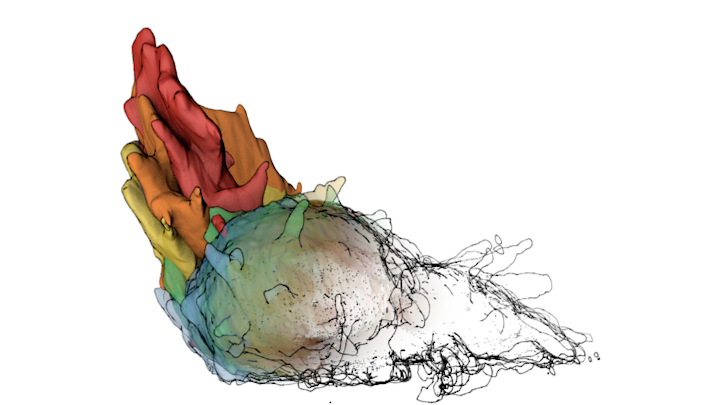
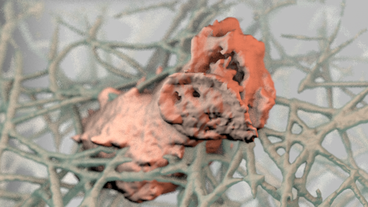
Megan Riel-Mehan The recently developed lattice light sheet microscope (LLSm) offers a unique combination of high spatial resolution, high speed and low phototoxicity. It is particularly well suited for visualizing fast moving, migrating cells such as white blood cells. However, the 3D time series data sets collected by the LLSm present many technical, visualization, and analysis challenges. Because this microscope is such a recent innovation, few existing analysis tools can visualize or process the data easily. In addition, there are limited visual conventions for exploring the data or communicating the results. The collaboration between the Mullins Cell Biology Lab (scientific visualization) and the Ferrin group (molecular viewing software) demonstrates how this shared project led to advances in our respective fields. Cell surfaces were imported into Cinema 4D then combined with more traditional illustration techniques of tonal shading and line to clarify surface topology and structural spatial relationships, among other complex analysis routines. The LLsm project exemplifies the many ways that visualization experts can help drive the scientific process and shows the benefits of engaging in collaboration early in the project timeline. I submitted 3 entries from my project with Dyche Mullins; you can vote every day. If you like any of these please vote for them.
Thanks! http://sci-resolution.strutta.com/entry/9767207 http://sci-resolution.strutta.com/entry/9767509 http://sci-resolution.strutta.com/entry/9767665 VOTING IS NOW OPEN for Sci-Resolution: UCSF’s Science Image & Video Competition! Who will win the People’s Choice award worth $500? 1. Register to vote at: http://sci-resolution.strutta.com/auth/login?destination=entries 2. Go to: http://sci-resolution.strutta.com/entries to view and vote for entries. 3. Vote for as many entries as you want once a day through October 15th
|

 RSS Feed
RSS Feed